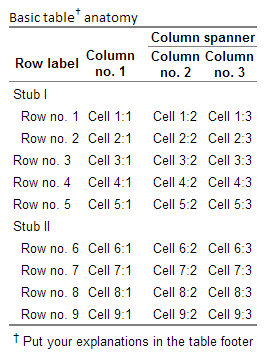
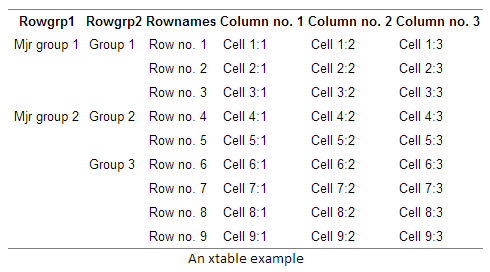
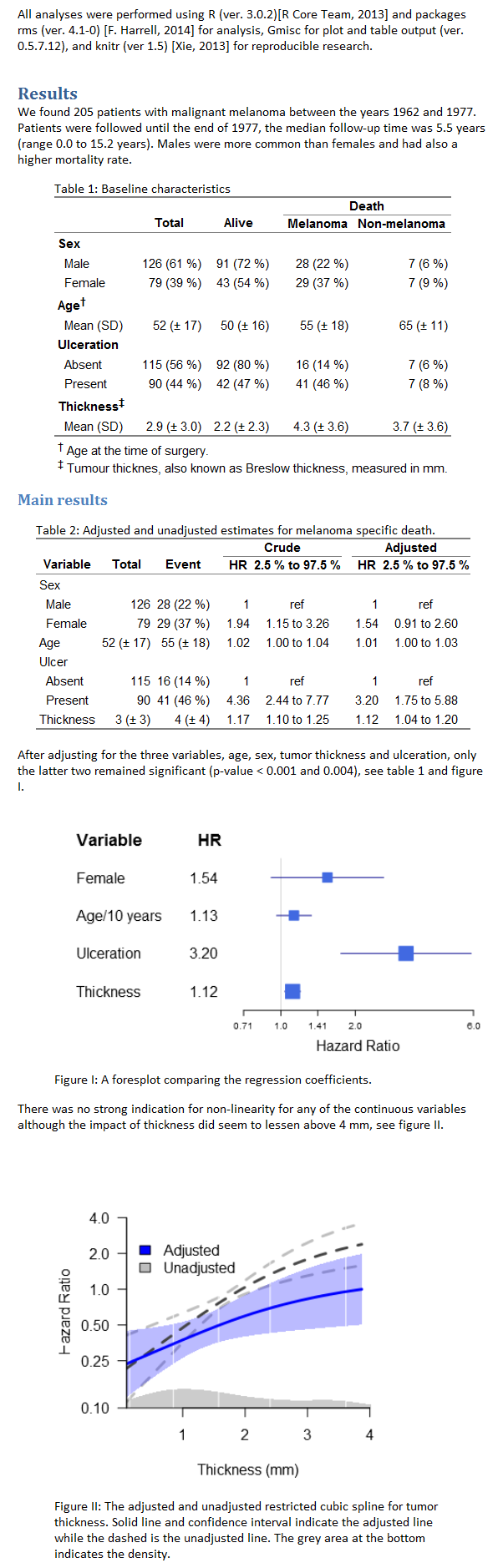
Fast-track publishing using knitr is a short series on how I use knitr to get my articles faster published. By fast-track publishing I mean eliminating as many of the obstacles as possible during the manuscript phase, and not the fast-track some journals offer. In this first introductory article I will try to (1) define the main groups of obstacles I have experienced motivating knitr, (2) options I’ve used for extracting knitted results into MS Word. The emphasis will be on general principles and what have worked for me.
Note: this introductory article contains no R-code. The next article in this series will show how to set-up a “MS Word-friendly” knitr markdown environment in RStudio and more (yes, there will be R-code).
Publishing obstacles
You (yes you) – in a stroke of brilliance, will realize that you want to re-categorize age into three groups instead of four… Keeping everything in knitr allows you to do this and more without even thinking twice.
Another common obstacle are your co-authors, these awesome people that unfortunately have a ton of useful input. Their input should in theory appear early in the process but are almost always late. A typical scenario is for instance that one of your co-authors realizes that you should exclude a few patients/observations, causing a chain-reaction in your document where you need to look through every table cell, every estimate and confidence interval, and every p-value. Having all these numbers auto-updating is therefore vital – a philosophy that is the foundation of knitr and its peers.
Once you’ve reached the phase of submitting your manuscript you will notice all the obstacles that the journal puts up. The main hint here is to look through the guidelines early on. If you’re uncertain what journal you’re submitting to, then start by reading the ICMJE preparing for submission guidelines that many follow. It is also good to follow either the CONSORT (RCT), PRISMA/QUORUM (meta-analysis), MOOSE (meta-analysis for observational studies), or STROBE (observational studies) statements. These guidelines are not journal specific, and following them makes switching journals simpler. Adhering to the statements will also make your article look more professional and reviewers will be faster in their response as they will be familiar with your document structure. When it comes to document demands, I’ve found a lot of journals to have two things in common: (1) Word-documents are always accepted (or at least PDF-converted), (2) as are TIFF/EPS(/JPEG) images. Adapting to Word format will make your life easier, while images are easily handled using knitr’s fig./dpi options.
The last and most important obstacle is reviewers’ input after submission – while often extensive (at least in my cases), addressing these will improve your article, and open the gates of publication. The worst part with these changes is that by the time you receive them you will have forgotten everything about your article. I cannot thank Yihui Xie and his predecessors/peers enough for making knitr that takes most of the pain out of the process. Knowing exactly where every number originates from, how you selected your final population etc. is gold when you try to go back into your masterpiece.
Options for getting text into Word
I have accepted the fact that my co-authors love Word and I therefore need to copy the text into a Word document as quickly as possible from markdown. There are a few options available that I’ve tried at one time or another, here’s the list and my experience with each and every one:
- Copying HTML-document from a browser: This works actually OK with MS Word (ver. 2010) if you have the proper formatting. Unfortunately it seems to change the set font-size for paragraphs from 11 to 11.5 – an annoying feature that can cause unwanted effects. Libre Office (ver. 4.1.4), in turn, fails with basics such as retaining bold <strong>, paragraph margins etc.
- Opening an HTML-document in a word processor: This my current method of choice. The only caveat is that formatted tables are generally ruined by MS Word and I therefore strongly recommend going through Libre Office. Libre Office is free, open source, and could be my primary editor if it wasn’t for my co-authors extreme love of Word. My current workflow is Markdown > Libre Office > Word. It may seem like a strange way of doing it, but it is reliable and sufficient.
- Using a markdown converter such as Pandoc: I have tried this once and it worked beautifully with the only exception that none of the HTML-tables worked. Markdown has its own limited table syntax, but since I find it insufficient I rely on HTML, and currently Pandoc does not support this when converting to Word. This is not that surprising as the layout options are endless for plain HTML-tables and building support for this is difficult. I hope that Pandoc in the future will support HTML-tables but until that time, it’s a deal breaker for me.
- Using R2DOCX: This is an awesome alternative but as far as I’m aware of there is no available RStudio integration, the table layout is not as flexible as HTML, and it’s still early in its development cycle. If you need to generate reports using a specific Word template, and writing is minimal this can be a good choice.
- Converting LaTeX-documents: I love LaTeX but converting it to Word is far from smooth. LaTeX is also bad at handling tables; it is great for text and formulas but the table solution is terrible – I’ve had countless documents fail to compile due to parts of a table being too wide.
- Using Google Docs: I’ve tried both copy-pasting and opening the document directly in Google Docs with very poor results. The table layout is completely lost in process.
Summary
Using knitr will give you a better connection between the statistics and the actual manuscript, thereby: (1) allowing late changes, (2) reproducing your results on demand, and (3) easier porting to final publishing format. Furthermore, getting more advanced formatted knitr documents into MS Word can be challenging, although with a little help from Libre Office this is not that difficult.
In next few posts I will cover above and show how to, avoid dense, poorly formatted manuscripts, and to use the page/table layout to your advantage – reviewers should find your article visually appealing and easy to read. Even if the journal remakes everything prior to publications, the reviewers (and the editor) are the ones you need make happy to get published.
R-bloggers.com offers daily e-mail updates about R news and tutorials on topics such as: visualization (ggplot2, Boxplots, maps, animation), programming (RStudio, Sweave, LaTeX, SQL, Eclipse, git, hadoop, Web Scraping) statistics (regression, PCA, time series, trading) and more...